Research
Protein-protein interactions in the Gram-negative bacterial cell envelope
In my laboratory, we aim to understand how protein-protein interactions in bacteria help support the organisation, structure and stability of the outer membrane and how such interactions can also be exploited by protein antibiotics known as bacteriocins to kill bacterial cells. Our research typically incorporates a range of biochemical, biophysical, structural and cell-based techniques.
Research Areas
Bacteriocins are species-specific protein antibiotics that parasitize a variety of outer membrane and periplasmic proteins in Gram-negative bacteria. Our work centres on the entry mechanisms of colicins, which target Escherichia coli, pyocins, which target Pseudomonas aeruginosa and klebicins, which target Klebsiella pneumoniae. These toxins serve as important agents of competition within microbial communities. We study both nuclease (DNases, rRNases and tRNases) and pore-forming bacteriocins, which use their network of protein-protein interactions within the cell envelope to establish a translocon complex that delivers a toxic domain into the cell. Hence, bacteriocin translocation represents a highly simplified model for understanding energised cellular protein import. We recently demonstrated that Tol- and Ton- dependent bacteriocins translocate across the outer membrane by the same basic mechanism in which the unfolded toxin is pulled through a specific translocator protein and powered by the proton-motive force of the inner membrane.
Porin threading drives receptor disengagement and establishes active colicin E9 transport through Escherichia coli OmpF. a-e, sequence of events at the E. coli cell surface as BtuB-bound ColE9 threads its disorder domain through two of the pores of OmpF. Threading causes the toxin to disengage from its receptor BtuB and associate with the periplasmic protein TolB. ColE9-TolB is dragged unfolded through a single subunit of OmpF by the PMF-linked inner membrane complex of TolQ-TolR-TolA. From the periplasm, ColE9 translocates across the inner membrane to deliver its cytotoxic nuclease (not shown). See Francis et al (2021) EMBO J and Housden et al (2013) Science for further details.
Tol-Pal is a little understood complex that is required for the stable maintenance of the Gram-negative outer membrane and which is recruited to the cell division apparatus late during division. We discovered a unique ‘mobilisation-and-capture’ mechanism underpins Tol-Pal function whereby force transduction across the cell envelope enables the accumulation of the peptidoglycan-binding protein Pal at division septa, stabilising the invaginating outer membrane. More recently, we solved the cryo-electron microscopy structure for the TolQR stator complex and demonstrated that structural rigidity within the force transducer protein TolA maximises delivery of mechanical force to the outer membrane following association with this PMF-coupled motor.
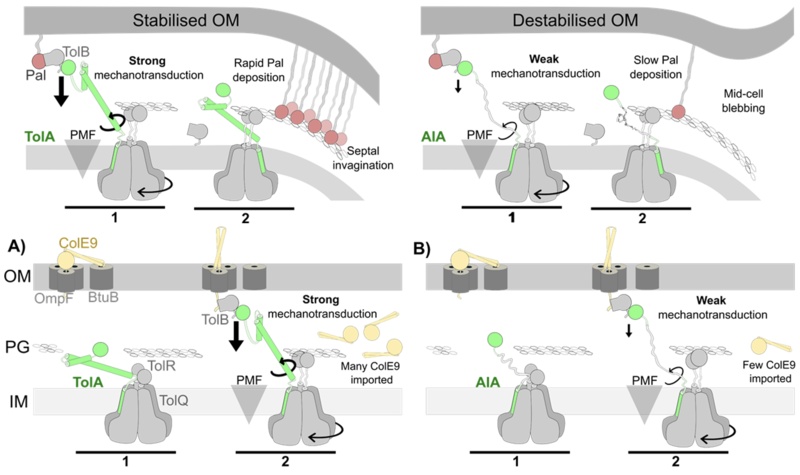
Efficient mechanotransduction of force by TolA to the outer membrane requires secondary structure in its periplasm spanning domain. Tol-Pal is a ubiquitous five-protein assembly in Gram-negative bacteria; TolQ, TolR and TolA are inner membrane proteins, Pal is an outer membrane lipoprotein and TolB is a periplasmic protein that binds Pal. The inner membrane complex of TolQ-TolR-TolA exploits the proton motive force (PMF) to dissociate the TolB-Pal complex, releasing Pal to bind the cell wall. During cell division the inner membrane complex becomes localised to the divisome enabling Pal deposition to also be localised. Increased [Pal] at the septum stabilises the invaginating outer membrane of the dividing cell. See Szczepaniak et al (2020) Nat Commun and Williams-Jones et al (2023) PNAS for further details.
The rise of multidrug resistant bacteria coupled with the lack of new classes of antibiotics in over 40 years means there is a pressing and urgent need for new antibiotics especially for molecules that target pathogenic Gram-negative bacteria. In collaboration with Prof Dan Walker at the University of Strathclyde, we have developed novel bacteriocin engineering pipelines that help avoid some of the traditional problems associated with using these potent toxins as antibiotics. These advances are the basis for a new spinout company, Glox Therapeutics Ltd, which is led by our CEO Dr James Clark.
Check out what Glox is doing at the company website: https://gloxtherapeutics.com/about/

Klebicin C exploits the drug efflux channel TolC to translocate across the outer membrane of Klebsiella pneumoniae. Klebicin C is a 60 kDa nuclease bacteriocin that specifically kills K. pneumoniae and closely related species. Top panel, 1.9 Å crystal structure of the 25 kDa N-terminal helical hairpin domain of Klebicin C. The hairpin opens like a switchblade knife to enter and bind TolC in the outer membrane. Lower panel, 3.1 Å cryo-electron microscopy structure of the Klebicin C-TolC complex. Following delivery of the bacteriocin’s TonB box (not shown) through the iris of TolC to the periplasm the entire toxin is dragged unfolded into the cell thereby delivering its nuclease payload. See Housden et al (2021) Nat Commun 12, 4625 for further details.
The asymmetric bilayer of the outer membrane contains outer membrane proteins (OMPs) and lipoproteins. Using colicins as OMP-specific probes, we discovered that OMPs cluster to form ‘OMP islands’ in the membrane and that this clustering behaviour underpins their turnover. Through the creation of a variety of OMP-specific labels we are dissecting the organisation of OMP islands and investigating the ramifications of these micro-domains to bacterial physiology. For example, we demonstrated that the inherent organisation of outer membrane proteins into islands becomes imprinted on inner membrane proteins when they are connected by the energized protein bridges. We also discovered that OMP clustering is mediated by interfacial LPS, creating an OMP-LPS-OMP network that spans the cell surface. The network is likely responsible for many of the outer membrane’s biophysical properties, such as its impermeability and rigidity.
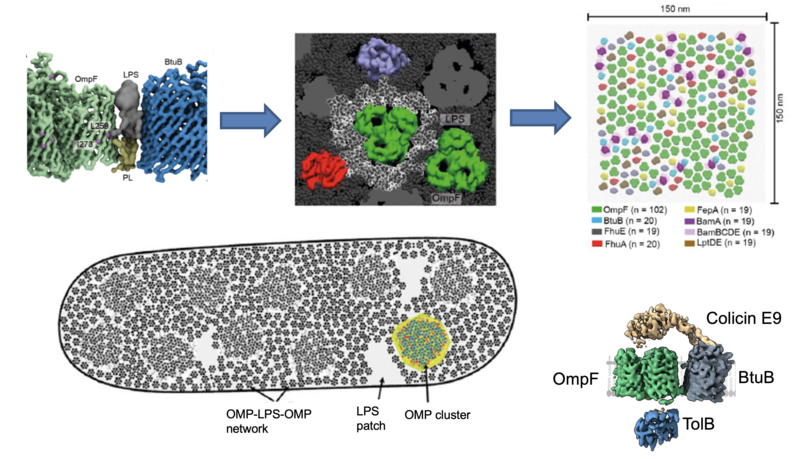
The E. coli outer membrane is a densely packed network of OMP-LPS-OMP assemblies. Near-neighbour, transmembrane crosslinking using the photoactivatable crosslinker BPA revealed that OMPs such as the trimeric porin OmpF are surrounded by a shell of asymmetric lipids. The same lipids mediate higher order assemblies with other OMPs leading to the formation of a cell-wide network of OMP-LPS-OMP complexes. Upper panels are MD snapshots (courtesy of Dheeraj Prakaash & Syma Khalid) of this basic unit of OM organisation, which results in a dense array of OMPs (centred on OmpF) with six-fold and three-fold symmetry. See Webby et al (2022) Sci Adv 8, eadc9566 for details. The OMP-LPS-OMP network restricts lateral mobility in the OM and explains, for example, how bacteriocins are able to ‘find’ two adjacent OMPs to trigger import into E. coli (the OM translocon structure for ColE9 is shown; see Francis et al (2021) EMBO J e108610 for details).
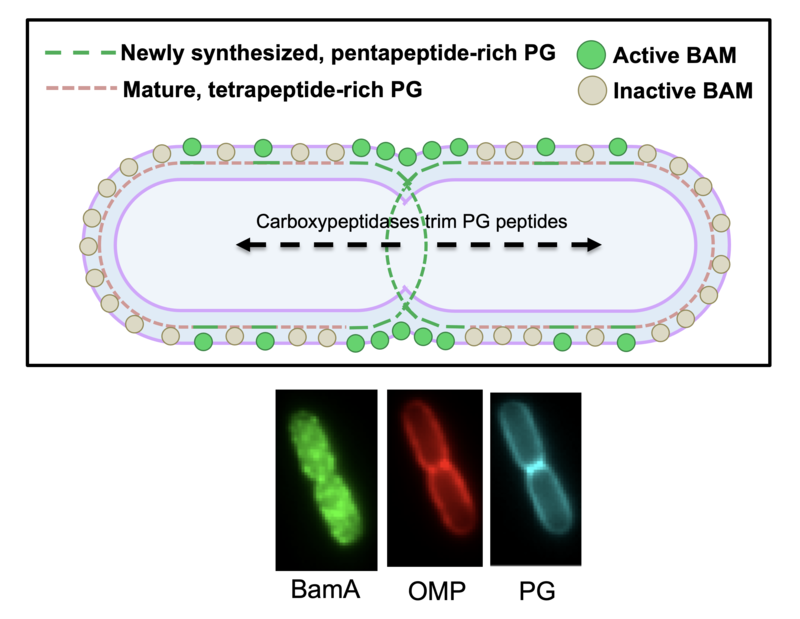
Peptidoglycan governs the spatiotemporal organisation of OMPs. In collaboration with Federico Corona and Waldemar Vollmer, we demonstrated that the maturation state of cell wall peptidoglycan is a major determinant of OMP insertion activity of the OMP insertase BamA. ‘Old’ PG, enriched at the old poles of dividing cells, contains predominantly tetrapeptide stem peptides that inhibit BamA activity. By contrast, ‘new’ PG at the division site, which is enriched in pentapeptide stem peptides, is a poor inhibitor of BamA. As a result, OMPs are inserted preferentially at division sites. Coupled with the lack of lateral mobility in the OM due to OMP-LPS-OMP networks, these observations explain how OMPs are turned over by binary partitioning and demonstrate that growth of PG and the OM is coordinated. See Rassam et al (2015) Nature 523, 333 and Mamou et al (2022) Nature 606, 953 for further details.


